Welcome to the kissim documentation
Subpocket-based structural fingerprint for kinase pocket comparison


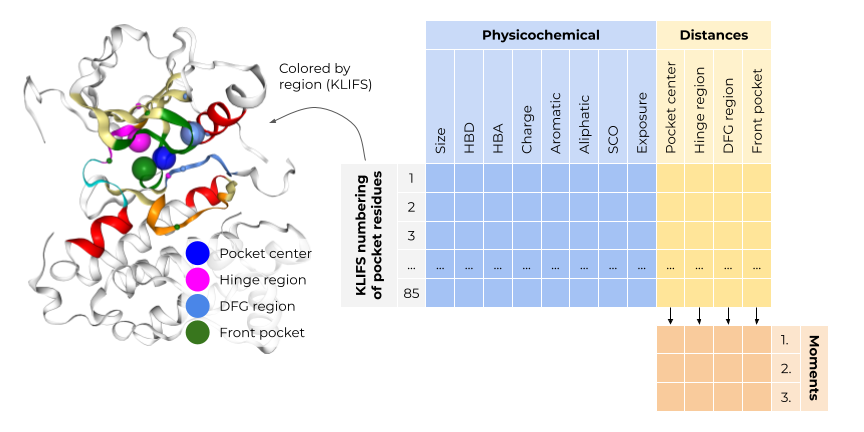
Subpocket-based structural fingerprint for kinase pockets
The kissim package offers a novel fingerprinting strategy designed specifically for kinase pockets,
allowing for similarity studies across the structurally covered kinome.
The kinase fingerprint is based on the KLIFS pocket alignment,
which defines 85 pocket residues for all kinase structures.
This enables a residue-by-residue comparison without a computationally expensive alignment step.
The pocket fingerprint consists of 85 concatenated residue fingerprints, each encoding a residue’s spatial and physicochemical properties. The spatial properties describe the residue’s position in relation to the kinase pocket center and important kinase subpockets, i.e. the hinge region, the DFG region, and the front pocket. The physicochemical properties encompass for each residue its size and pharmacophoric features, solvent exposure and side chain orientation.
Take a look at the kissim_app repository for pairwise comparison of all kinases to study kinome-wide similarities.
Explore package
Developers