API - Quick start Python interface
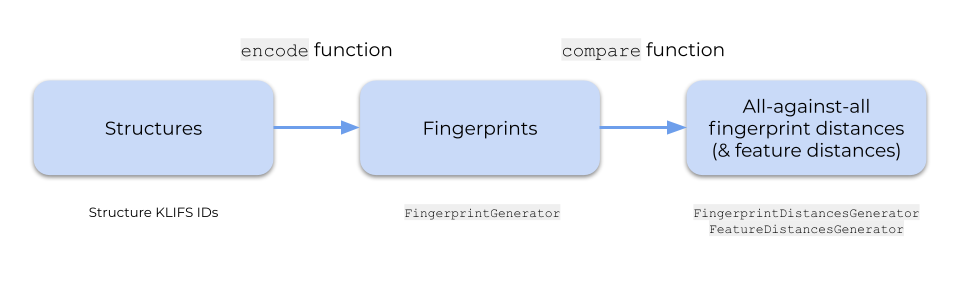
Let’s take a look at the kissim quick start API to encode a set of structures (from the KLIFS database) and perform an all-against-all comparison.
[1]:
from kissim.api import encode, compare
[2]:
# Load path to test data
from kissim.dataset.test import PATH as PATH_TEST_DATA
Encode structures into fingerprints
The encode function is a quick start API to generate fingerprints in bulk based on structure KLIFS IDs.
Input parameters are:
structure_klifs_ids: Structure KLIFS IDs.fingerprints_json_filepath: (Optionally) Path to output json file containing fingerprints.local_klifs_download_path: (Optionally) Set path local KLIFS download or - if not set - fetch data from KLIFS database.n_cores: (Optionally) Number of cores used to generate fingerprints.
The return object is of type FingerprintGenerator.
[3]:
# flake8-noqa-cell
encode?
Signature:
encode(
structure_klifs_ids,
fingerprints_filepath=None,
local_klifs_download_path=None,
n_cores=1,
)
Docstring:
Encode structures.
Parameters
----------
structure_klifs_ids : list of int
Structure KLIFS IDs.
fingerprints_filepath : str or pathlib.Path
Path to output json file. Default None.
local_klifs_download_path : str or None
If path to local KLIFS download is given, set up local KLIFS session.
If None is given, set up remote KLIFS session.
n_cores : int
Number of cores used to generate fingerprints.
Returns
-------
kissim.encoding.FingerprintGenerator
Fingerprints.
File: ~/Documents/GitHub/kissim/kissim/api/encode.py
Type: function
Run encode function
[4]:
structure_klifs_ids = [109, 118, 12347, 1641, 3833, 9122]
fingerprint_generator = encode(
structure_klifs_ids=structure_klifs_ids,
fingerprints_filepath=None,
n_cores=2,
local_klifs_download_path=PATH_TEST_DATA / "KLIFS_download",
)
Inspect output: FingerprintGenerator
[5]:
print(f"Number of structures (input): {len(structure_klifs_ids)}")
print(f"Number of fingerprints (output): {len(fingerprint_generator.data.keys())}")
fingerprint_generator
Number of structures (input): 6
Number of fingerprints (output): 6
[5]:
<kissim.encoding.fingerprint_generator.FingerprintGenerator at 0x7f04c1976910>
[6]:
fingerprint_generator.data
[6]:
{109: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18d0970>,
118: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18dd1c0>,
12347: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18d0b80>,
1641: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18d03a0>,
3833: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18d0430>,
9122: <kissim.encoding.fingerprint.Fingerprint at 0x7f04c18dd130>}
Find more information about the FingerprintGenerator object here.
Compare fingerprints
The compare function is a quick start API to perform a pairwise all-against-all (bulk) comparison for a set of fingerprints.
Input parameters are:
fingerprint_generator: Fingerprints.output_path: (Optionally) Path to output folder for distances json files.feature_weights: (Optionally) Feature weights used to calculate the final fingerprint distance.n_cores: (Optionally) Number of cores used to generate distances.
The return objects are of type FeatureDistancesGenerator and FingerprintDistanceGenerator.
[7]:
# flake8-noqa-cell
compare?
Signature:
compare(
fingerprint_generator,
output_path=None,
feature_weights=None,
n_cores=1,
)
Docstring:
Compare fingerprints (pairwise).
Parameters
----------
fingerprint_generator : kissim.encoding.FingerprintGenerator
Fingerprints for KLIFS dataset.
output_path : str
Path to output folder.
feature_weights : None or list of float
Feature weights of the following form:
(i) None
Default feature weights: All features equally distributed to 1/15
(15 features in total).
(ii) By feature (list of 15 floats):
Features to be set in the following order: size, hbd, hba, charge, aromatic,
aliphatic, sco, exposure, distance_to_centroid, distance_to_hinge_region,
distance_to_dfg_region, distance_to_front_pocket, moment1, moment2, and moment3.
All floats must sum up to 1.0.
n_cores : int
Number of cores used to generate fingerprint distances.
Returns
-------
feature_distances_generator : kissim.comparison.FeatureDistancesGenerator
Feature distances for all pairwise fingerprints.
fingerprint_distance_generator : kissim.comparison.FingerprintDistanceGenerator
Fingerprint distance for all pairwise fingeprints.
File: ~/Documents/GitHub/kissim/kissim/api/compare.py
Type: function
Run compare function
[8]:
feature_distances_generator, fingerprint_distance_generator = compare(
fingerprint_generator=fingerprint_generator,
output_path=None,
feature_weights=None,
n_cores=2,
)
For final fingerprint distances, please refer to the FingerprintDistanceGenerator object.
Inspect output: FingerprintDistanceGenerator
[9]:
print(f"Number of fingerprints (input): {len(fingerprint_generator.data)}")
print(f"Number of pairwise comparisons (output): {len(fingerprint_distance_generator.data)}")
fingerprint_distance_generator
Number of fingerprints (input): 6
Number of pairwise comparisons (output): 15
[9]:
<kissim.comparison.fingerprint_distance_generator.FingerprintDistanceGenerator at 0x7f04c1880310>
[10]:
fingerprint_distance_generator.data
[10]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance | bit_coverage | |
|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.074214 | 0.992000 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.259053 | 0.919333 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.253045 | 0.990667 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.277368 | 0.990667 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.358882 | 0.990667 |
| 5 | 118 | 12347 | ABL2 | BRAF | 0.273133 | 0.918000 |
| 6 | 118 | 1641 | ABL2 | CHK1 | 0.246844 | 0.989333 |
| 7 | 118 | 3833 | ABL2 | AAK1 | 0.282949 | 0.990000 |
| 8 | 118 | 9122 | ABL2 | ADCK3 | 0.360833 | 0.989333 |
| 9 | 12347 | 1641 | BRAF | CHK1 | 0.303330 | 0.918000 |
| 10 | 12347 | 3833 | BRAF | AAK1 | 0.307277 | 0.918000 |
| 11 | 12347 | 9122 | BRAF | ADCK3 | 0.376875 | 0.918000 |
| 12 | 1641 | 3833 | CHK1 | AAK1 | 0.229590 | 0.989333 |
| 13 | 1641 | 9122 | CHK1 | ADCK3 | 0.347142 | 0.989333 |
| 14 | 3833 | 9122 | AAK1 | ADCK3 | 0.303542 | 0.990000 |
Get the structure distance matrix.
[11]:
fingerprint_distance_generator.structure_distance_matrix()
[11]:
| structure.2 | 109 | 118 | 1641 | 3833 | 9122 | 12347 |
|---|---|---|---|---|---|---|
| structure.1 | ||||||
| 109 | 0.000000 | 0.074214 | 0.253045 | 0.277368 | 0.358882 | 0.259053 |
| 118 | 0.074214 | 0.000000 | 0.246844 | 0.282949 | 0.360833 | 0.273133 |
| 1641 | 0.253045 | 0.246844 | 0.000000 | 0.229590 | 0.347142 | 0.303330 |
| 3833 | 0.277368 | 0.282949 | 0.229590 | 0.000000 | 0.303542 | 0.307277 |
| 9122 | 0.358882 | 0.360833 | 0.347142 | 0.303542 | 0.000000 | 0.376875 |
| 12347 | 0.259053 | 0.273133 | 0.303330 | 0.307277 | 0.376875 | 0.000000 |
Map structure pairs to kinase pairs (example: here use structure pair with minimum distance as representative for kinase pair).
[12]:
fingerprint_distance_generator.kinase_distance_matrix(by="minimum")
[12]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | BRAF | CHK1 |
|---|---|---|---|---|---|
| kinase.1 | |||||
| AAK1 | 0.000000 | 0.277368 | 0.303542 | 0.307277 | 0.229590 |
| ABL2 | 0.277368 | 0.000000 | 0.358882 | 0.259053 | 0.246844 |
| ADCK3 | 0.303542 | 0.358882 | 0.000000 | 0.376875 | 0.347142 |
| BRAF | 0.307277 | 0.259053 | 0.376875 | 0.000000 | 0.303330 |
| CHK1 | 0.229590 | 0.246844 | 0.347142 | 0.303330 | 0.000000 |
Inspect output: FeatureDistancesGenerator
[13]:
print(f"Number of fingerprints (input): {len(fingerprint_generator.data.keys())}")
print(f"Number of pairwise comparisons (output): {len(feature_distances_generator.data)}")
feature_distances_generator
Number of fingerprints (input): 6
Number of pairwise comparisons (output): 15
[13]:
<kissim.comparison.feature_distances_generator.FeatureDistancesGenerator at 0x7f0524591ee0>
[14]:
feature_distances_generator.data
[14]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance.1 | distance.2 | distance.3 | distance.4 | distance.5 | distance.6 | ... | bit_coverage.6 | bit_coverage.7 | bit_coverage.8 | bit_coverage.9 | bit_coverage.10 | bit_coverage.11 | bit_coverage.12 | bit_coverage.13 | bit_coverage.14 | bit_coverage.15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.243590 | 0.141026 | 0.230769 | ... | 0.92 | 0.67 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.247059 | 0.141176 | 0.223529 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.211765 | 0.082353 | 0.270588 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.258824 | 0.235294 | 0.305882 | ... | 1.00 | 0.88 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 5 | 118 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.243590 | 0.141026 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 6 | 118 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.247059 | 0.141176 | 0.223529 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 7 | 118 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.211765 | 0.082353 | 0.270588 | ... | 1.00 | 0.85 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 8 | 118 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.258824 | 0.235294 | 0.305882 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 9 | 12347 | 1641 | BRAF | CHK1 | 0.346154 | 0.423077 | 0.346154 | 0.243590 | 0.115385 | 0.217949 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 10 | 12347 | 3833 | BRAF | AAK1 | 0.435897 | 0.474359 | 0.333333 | 0.230769 | 0.102564 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 11 | 12347 | 9122 | BRAF | ADCK3 | 0.576923 | 0.371795 | 0.397436 | 0.256410 | 0.269231 | 0.282051 | ... | 0.92 | 0.68 | 0.89 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 12 | 1641 | 3833 | CHK1 | AAK1 | 0.352941 | 0.411765 | 0.352941 | 0.200000 | 0.082353 | 0.235294 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 13 | 1641 | 9122 | CHK1 | ADCK3 | 0.611765 | 0.494118 | 0.541176 | 0.317647 | 0.282353 | 0.317647 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 14 | 3833 | 9122 | AAK1 | ADCK3 | 0.611765 | 0.482353 | 0.494118 | 0.282353 | 0.223529 | 0.341176 | ... | 1.00 | 0.87 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
15 rows × 34 columns