CLI - Command-line interface
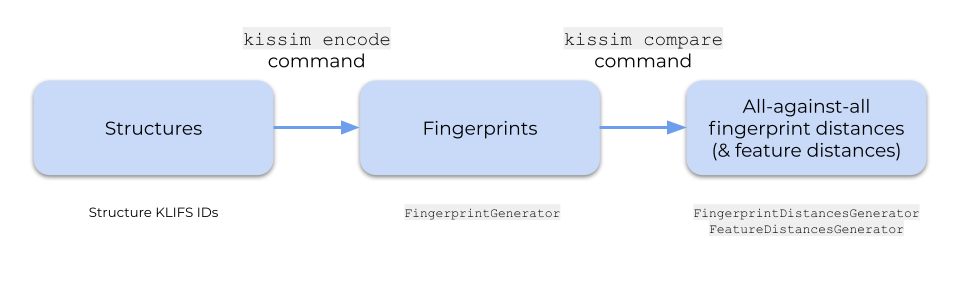
Let’s take a look at the kissim command-line interface (CLI) to encode a set of structures (from the KLIFS database) and perform an all-against-all comparison. The CLI follows the same logic as the quick start Python interface as described in “API - Quick start Python interface”.
[1]:
from pathlib import Path
import pandas as pd
[2]:
# Path to this notebook
HERE = Path(_dh[-1]) # noqa: F821
Encode structures into fingerprints
[3]:
%%bash
kissim encode -h
# flake8-noqa-cell
usage: kissim encode [-h] -i INPUT [INPUT ...] -o OUTPUT [-l LOCAL]
[-c NCORES]
optional arguments:
-h, --help show this help message and exit
-i INPUT [INPUT ...], --input INPUT [INPUT ...]
List of structure KLIFS IDs or path to txt file
containing structure KLIFS IDs.
-o OUTPUT, --output OUTPUT
Path to output JSON file containing fingerprint data.
-l LOCAL, --local LOCAL
Path to KLIFS download folder. If set local KLIFS data
is used, else remote KLIFS data.
-c NCORES, --ncores NCORES
Number of cores. If 1 fingerprint generation in
sequence, else in parallel.
kissim encode command
[4]:
%%bash
# Set path to local KLIFS data
PATH_KLIFS_DOWNLOAD="../../kissim/tests/data/KLIFS_download/"
kissim encode -i 109 118 12347 1641 3833 9122 -o fingerprints.json -l $PATH_KLIFS_DOWNLOAD -c 2
# flake8-noqa-cell
INFO:kissim.encoding.fingerprint_generator:GENERATE FINGERPRINTS
INFO:kissim.encoding.fingerprint_generator:Number of input structures: 6
INFO:kissim.encoding.fingerprint_generator:Fingerprint generation started at: 2021-11-16 14:21:15.937623
INFO:kissim.utils:Number of cores used: 2.
INFO:kissim.encoding.fingerprint_generator:109: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:118: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:12347: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:1641: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:3833: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:9122: Generate fingerprint...
INFO:kissim.encoding.fingerprint_generator:Number of output fingerprints: 6
INFO:kissim.encoding.fingerprint_generator:Runtime: 0:00:06.288938
INFO:kissim.api.encode:Write fingerprints to file: fingerprints.json
This command generate two files:
Data:
fingerprints.jsonLogs (not under Windows):
fingerprint.log
Inspect output: FingerprintGenerator
You can load the content of the fingerprints.json file as FingerprintGenerator object.
[5]:
fingerprints_path = HERE / "fingerprints.json"
fingerprints_path
[5]:
PosixPath('/home/dominique/Documents/GitHub/kissim/docs/tutorials/fingerprints.json')
[6]:
from kissim.encoding import FingerprintGenerator
fingerprint_generator = FingerprintGenerator.from_json(fingerprints_path)
print(f"Number of fingerprints: {len(fingerprint_generator.data.keys())}")
fingerprint_generator
Number of fingerprints: 6
[6]:
<kissim.encoding.fingerprint_generator.FingerprintGenerator at 0x7f78c6d10ca0>
Compare fingerprints
[7]:
%%bash
kissim compare -h
# flake8-noqa-cell
usage: kissim compare [-h] -i INPUT -o OUTPUT
[-w WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS]
[-c NCORES]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Path to JSON file containing fingerprint data.
-o OUTPUT, --output OUTPUT
Path to output folder where distance bzip-compressed
CSV files will be saved.
-w WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS, --weights WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS WEIGHTS
Feature weights. Eeach feature must be set
individually, all weights must sum up to 1.0.
-c NCORES, --ncores NCORES
Number of cores. If 1 comparison in sequence, else in
parallel.
kissim compare command
[8]:
%%bash
kissim compare -i fingerprints.json -o . -c 2
# flake8-noqa-cell
INFO:kissim.comparison.feature_distances_generator:GENERATE FEATURE DISTANCES
INFO:kissim.comparison.feature_distances_generator:Number of input fingerprints: 6
INFO:kissim.comparison.feature_distances_generator:Feature distances generation started at: 2021-11-16 14:21:25.566224
INFO:kissim.utils:Number of cores used: 2.
INFO:kissim.comparison.feature_distances_generator:Number of ouput feature distances: 15
INFO:kissim.comparison.feature_distances_generator:Runtime: 0:00:00.058451
INFO:kissim.comparison.fingerprint_distance_generator:GENERATE FINGERPRINT DISTANCES
INFO:kissim.comparison.fingerprint_distance_generator:Fingerprint distance generation started at: 2021-11-16 14:21:25.627493
INFO:kissim.comparison.fingerprint_distance_generator:Feature weights: [0.06666667 0.06666667 0.06666667 0.06666667 0.06666667 0.06666667
0.06666667 0.06666667 0.06666667 0.06666667 0.06666667 0.06666667
0.06666667 0.06666667 0.06666667]
Calculate pairwise fingerprint distance: 100%|██████████| 15/15 [00:00<00:00, 22614.87it/s]
Calculate pairwise fingerprint coverage: 100%|██████████| 15/15 [00:00<00:00, 25115.59it/s]
INFO:kissim.comparison.fingerprint_distance_generator:Number of output fingerprint distances: 15
INFO:kissim.comparison.fingerprint_distance_generator:Runtime: 0:00:00.004275
INFO:kissim.comparison.tree:Clustering (method: ward) and calculating branch distances
INFO:kissim.comparison.tree:Converting clustering to a Newick tree
INFO:kissim.comparison.tree:Writing resulting tree to fingerprint_distances_to_kinase_clusters.tree
INFO:kissim.comparison.tree:Writing resulting kinase annotation to kinase_annotation.csv
This command generate the following files:
Data - fingerprint distances:
fingerprint_distances.csv.bz2Data - feature distances:
feature_distances.csv.bz2Data - default kinase matrix based on minimum fingerprint pair distance per kinase pair:
fingerprint_distances_to_kinase_matrix.csvData - default kinase tree based on kinase matrix clustering (hierarchical clustering using Ward’s method):
fingerprint_distances_to_kinase_clusters.csvLogs (not under Windows):
distances.log
Inspect output: All-against-all fingerprint distances
You can load the content of the fingerprint_distances.csv.bz2 file as FingerprintDistancesGenerator object.
[9]:
from kissim.comparison import FingerprintDistanceGenerator
fingerprint_distance_path = HERE / "fingerprint_distances.csv.bz2"
fingerprint_distance_generator = FingerprintDistanceGenerator.from_csv(fingerprint_distance_path)
print(f"Number of pairwise comparisons: {len(fingerprint_distance_generator.data)}")
fingerprint_distance_generator
Number of pairwise comparisons: 15
[9]:
<kissim.comparison.fingerprint_distance_generator.FingerprintDistanceGenerator at 0x7f78c01baa90>
[10]:
fingerprint_distance_generator.data
[10]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance | bit_coverage | |
|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.074214 | 0.992000 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.256488 | 0.919333 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.251476 | 0.990667 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.278152 | 0.990667 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.357313 | 0.990667 |
| 5 | 118 | 12347 | ABL2 | BRAF | 0.270569 | 0.918000 |
| 6 | 118 | 1641 | ABL2 | CHK1 | 0.245275 | 0.989333 |
| 7 | 118 | 3833 | ABL2 | AAK1 | 0.283733 | 0.990000 |
| 8 | 118 | 9122 | ABL2 | ADCK3 | 0.359265 | 0.989333 |
| 9 | 12347 | 1641 | BRAF | CHK1 | 0.300766 | 0.918000 |
| 10 | 12347 | 3833 | BRAF | AAK1 | 0.305568 | 0.918000 |
| 11 | 12347 | 9122 | BRAF | ADCK3 | 0.375166 | 0.918000 |
| 12 | 1641 | 3833 | CHK1 | AAK1 | 0.227237 | 0.989333 |
| 13 | 1641 | 9122 | CHK1 | ADCK3 | 0.344004 | 0.989333 |
| 14 | 3833 | 9122 | AAK1 | ADCK3 | 0.301189 | 0.990000 |
Inspect output: All-against-all feature distances
You can load the content of the feature_distances.csv.bz2 file as FeatureDistancesGenerator object.
[11]:
from kissim.comparison import FeatureDistancesGenerator
feature_distances_path = HERE / "feature_distances.csv.bz2"
feature_distances_generator = FeatureDistancesGenerator.from_csv(feature_distances_path)
print(f"Number of pairwise comparisons: {len(feature_distances_generator.data)}")
feature_distances_generator
Number of pairwise comparisons: 15
[11]:
<kissim.comparison.feature_distances_generator.FeatureDistancesGenerator at 0x7f78c00c65b0>
[12]:
feature_distances_generator.data
[12]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance.1 | distance.2 | distance.3 | distance.4 | distance.5 | distance.6 | ... | bit_coverage.6 | bit_coverage.7 | bit_coverage.8 | bit_coverage.9 | bit_coverage.10 | bit_coverage.11 | bit_coverage.12 | bit_coverage.13 | bit_coverage.14 | bit_coverage.15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.205128 | 0.141026 | 0.230769 | ... | 0.92 | 0.67 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.223529 | 0.141176 | 0.223529 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.223529 | 0.082353 | 0.270588 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.235294 | 0.235294 | 0.305882 | ... | 1.00 | 0.88 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 5 | 118 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.205128 | 0.141026 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 6 | 118 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.223529 | 0.141176 | 0.223529 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 7 | 118 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.223529 | 0.082353 | 0.270588 | ... | 1.00 | 0.85 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 8 | 118 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.235294 | 0.235294 | 0.305882 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 9 | 12347 | 1641 | BRAF | CHK1 | 0.346154 | 0.423077 | 0.346154 | 0.205128 | 0.115385 | 0.217949 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 10 | 12347 | 3833 | BRAF | AAK1 | 0.435897 | 0.474359 | 0.333333 | 0.205128 | 0.102564 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 11 | 12347 | 9122 | BRAF | ADCK3 | 0.576923 | 0.371795 | 0.397436 | 0.230769 | 0.269231 | 0.282051 | ... | 0.92 | 0.68 | 0.89 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 12 | 1641 | 3833 | CHK1 | AAK1 | 0.352941 | 0.411765 | 0.352941 | 0.164706 | 0.082353 | 0.235294 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 13 | 1641 | 9122 | CHK1 | ADCK3 | 0.611765 | 0.494118 | 0.541176 | 0.270588 | 0.282353 | 0.317647 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 14 | 3833 | 9122 | AAK1 | ADCK3 | 0.611765 | 0.482353 | 0.494118 | 0.247059 | 0.223529 | 0.341176 | ... | 1.00 | 0.87 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
15 rows × 34 columns
Clean up output files.
Inspect output: Kinase matrix
[13]:
kinase_matrix = pd.read_csv(HERE / "fingerprint_distances_to_kinase_matrix.csv", index_col=0)
kinase_matrix
[13]:
| AAK1 | ABL2 | ADCK3 | BRAF | CHK1 | |
|---|---|---|---|---|---|
| AAK1 | 0.000000 | 0.278152 | 0.301189 | 0.305568 | 0.227237 |
| ABL2 | 0.278152 | 0.000000 | 0.357313 | 0.256488 | 0.245275 |
| ADCK3 | 0.301189 | 0.357313 | 0.000000 | 0.375166 | 0.344004 |
| BRAF | 0.305568 | 0.256488 | 0.375166 | 0.000000 | 0.300766 |
| CHK1 | 0.227237 | 0.245275 | 0.344004 | 0.300766 | 0.000000 |
Inspect output: Kinome tree and kinase annotation
[14]:
# Kinome tree
with open(HERE / "fingerprint_distances_to_kinase_clusters.tree", "r") as f:
newick_string = f.read()
print(newick_string)
(((BRAF:0.256,ABL2:0.256)0.256:0.063,(CHK1:0.227,AAK1:0.227)0.227:0.092)0.269:0.064,ADCK3:0.384);
[15]:
# Kinase annotation
kinase_annotations = pd.read_csv(HERE / "kinase_annotation.csv", index_col=0, sep="\t")
kinase_annotations
[15]:
| kinase.family | kinase.group | |
|---|---|---|
| kinase.klifs_name | ||
| CHK1 | CAMKL | CAMK |
| BRAF | RAF | TKL |
| ABL2 | Abl | TK |
| AAK1 | NAK | Other |
| ADCK3 | ABC1 | Atypical |
The Newick tree and kinase annotation files can be loaded into the external software FigTree to visualize the kissim-based kinome tree.
Delete output files
[16]:
[i.unlink() for i in HERE.glob("*.json")]
[i.unlink() for i in HERE.glob("*.csv")]
[i.unlink() for i in HERE.glob("*.csv.bz2")]
[i.unlink() for i in HERE.glob("*.tree")]
[i.unlink() for i in HERE.glob("*.log")];