Comparison
The distance between two fingerprints is calculated in two steps:
Calculate feature distances: Calculate distance for each feature (e.g. distance between pairwise 85 size feature bits). Use L1 norm for discrete features and L2 norm for continuous features (scaled by number of bits per feature).
Calculate fingerprint distance: Calculate the weighted sum of all feature distances (sum of feature weights equals 1).
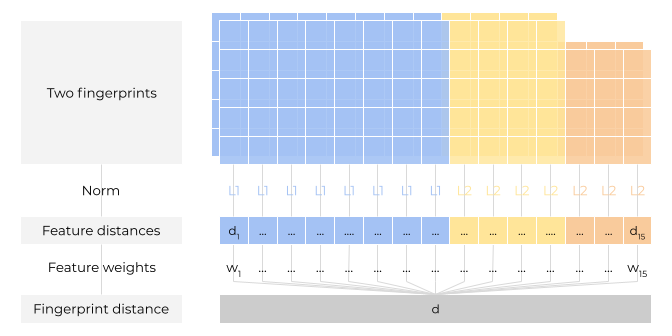
Figure 1: The pairwise kissim fingerprint comparison.
Respective objects performing these calculates are the FeatureDistances and FingerprintDistance objects. Furthermore, such distances can not only be generated between two fingerprints as described above but also in bulk for a set of fingerprints in an all-against-all comparison using the objects FingerprintDistanceGenerator and FeatureDistancesGenerator.
Let’s take a look at the API logic in this table again:
Action |
Module |
Single calculation |
Bulk calculation |
|---|---|---|---|
Encode structures as fingerprint |
|
|
|
Compare fingerprint features (calculate feature distances) |
|
|
|
Compare fingerprints (calculate fingerprint distance) |
|
|
|
[1]:
# Load path to test data
from kissim.dataset.test import PATH as PATH_TEST_DATA
Set up local KLIFS session using the opencadd.databases.klifs module.
[2]:
from opencadd.databases.klifs import setup_local
KLIFS_LOCAL = setup_local(PATH_TEST_DATA / "KLIFS_download")
Select structure KLIFS IDs
[3]:
structure_klifs_ids = [109, 118, 12347, 1641, 3833, 9122]
Generate fingerprints
Let’s generate a few fingerprints for the structures in our local KLIFS download using the bulk fingerprint generator FingerprintGenerator.
[4]:
from kissim.encoding import FingerprintGenerator
fingerprint_generator = FingerprintGenerator.from_structure_klifs_ids(
structure_klifs_ids=structure_klifs_ids, klifs_session=KLIFS_LOCAL
)
print(f"Number of fingerprints: {len(fingerprint_generator.data.keys())}")
Number of fingerprints: 6
Note: If fingerprint cannot be generated (e.g. because structural data is missing), the structure is skipped.
Compare two fingerprints
Let’s first focus on the comparison between two fingerprints only.
For two fingerprints (Fingerprint objects), we will
Calculate the feature distances using
FeatureDistancesandCalculate based on these feature distances and given feature weights the final fingerprint distance using
FingerprintDistance.
Generate feature distances between two fingerprints (FeatureDistances)
Input: Two
FingerprintobjectsOutput:
FeatureDistancesobject
[5]:
fingerprints = list(fingerprint_generator.data.values())
fingerprint1 = fingerprints[0]
fingerprint2 = fingerprints[1]
[6]:
from kissim.comparison import FeatureDistances
feature_distances = FeatureDistances.from_fingerprints(fingerprint1, fingerprint2)
print(f"Kinase pair: {feature_distances.kinase_pair_ids}")
print(f"Structure pair: {feature_distances.structure_pair_ids}")
feature_distances.data
Kinase pair: ('ABL2', 'ABL2')
Structure pair: (109, 118)
[6]:
| feature_type | feature_name | distance | bit_coverage | |
|---|---|---|---|---|
| 0 | physicochemical | size | 0.000000 | 1.00 |
| 1 | physicochemical | hbd | 0.000000 | 1.00 |
| 2 | physicochemical | hba | 0.000000 | 1.00 |
| 3 | physicochemical | charge | 0.000000 | 1.00 |
| 4 | physicochemical | aromatic | 0.000000 | 1.00 |
| 5 | physicochemical | aliphatic | 0.000000 | 1.00 |
| 6 | physicochemical | sco | 0.080000 | 0.88 |
| 7 | physicochemical | exposure | 0.294118 | 1.00 |
| 8 | distances | distance_to_centroid | 0.059839 | 1.00 |
| 9 | distances | distance_to_hinge_region | 0.122168 | 1.00 |
| 10 | distances | distance_to_dfg_region | 0.105499 | 1.00 |
| 11 | distances | distance_to_front_pocket | 0.070291 | 1.00 |
| 12 | moments | moment1 | 0.060816 | 1.00 |
| 13 | moments | moment2 | 0.116013 | 1.00 |
| 14 | moments | moment3 | 0.204469 | 1.00 |
Generate fingerprint distance between two fingerprints (FingerprintDistance)
Input:
FeatureDistancesobject and optionally feature weightsOutput:
FingerprintDistanceobject
Use standard feature weights
[7]:
from kissim.comparison import FingerprintDistance
fingerprint_distance = FingerprintDistance.from_feature_distances(
feature_distances, feature_weights=None
)
print(f"Fingerprint distance: {fingerprint_distance.distance}")
print(f"Fingerprint bit coverage: {fingerprint_distance.bit_coverage}")
Fingerprint distance: 0.07421423894307076
Fingerprint bit coverage: 0.9919999999999999
Use user-defined feature weights
[8]:
feature_weights = [0.3 / 8] * 8 + [0.5 / 4] * 4 + [0.2 / 3] * 3
fingerprint_distance = FingerprintDistance.from_feature_distances(
feature_distances, feature_weights=feature_weights
)
print(f"Fingerprint distance: {fingerprint_distance.distance}")
print(f"Fingerprint bit coverage: {fingerprint_distance.bit_coverage}")
Fingerprint distance: 0.08417398268335104
Fingerprint bit coverage: 0.9954999999999999
Compare all-against-all fingerprints
Let’s now take a look at the bulk distance generators to generate all-against-all comparisons for a set of fingerprints.
For a FingerprintGenerator object, which contains the fingerprints for a set of structures, we will
Calculate feature distances for all fingerprint pairs using
FeatureDistancesGeneratorandCalculate based on these feature distances and given feature weights the final fingerprint distance for all fingerprint pairs using
FingerprintDistanceGenerator.
Generate feature distances for all pairwise structures/fingerprints (FeatureDistancesGenerator)
Input:
FingerprintGeneratorobjectOutput:
FeatureDistancesGeneratorobject
[9]:
from kissim.comparison import FeatureDistancesGenerator
feature_distances_generator = FeatureDistancesGenerator.from_fingerprint_generator(
fingerprint_generator
)
feature_distances_generator.data
[9]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance.1 | distance.2 | distance.3 | distance.4 | distance.5 | distance.6 | ... | bit_coverage.6 | bit_coverage.7 | bit_coverage.8 | bit_coverage.9 | bit_coverage.10 | bit_coverage.11 | bit_coverage.12 | bit_coverage.13 | bit_coverage.14 | bit_coverage.15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.243590 | 0.141026 | 0.230769 | ... | 0.92 | 0.67 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.247059 | 0.141176 | 0.223529 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.211765 | 0.082353 | 0.270588 | ... | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.258824 | 0.235294 | 0.305882 | ... | 1.00 | 0.88 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 5 | 118 | 12347 | ABL2 | BRAF | 0.410256 | 0.397436 | 0.333333 | 0.243590 | 0.141026 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 6 | 118 | 1641 | ABL2 | CHK1 | 0.388235 | 0.352941 | 0.364706 | 0.247059 | 0.141176 | 0.223529 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 7 | 118 | 3833 | ABL2 | AAK1 | 0.505882 | 0.505882 | 0.411765 | 0.211765 | 0.082353 | 0.270588 | ... | 1.00 | 0.85 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 8 | 118 | 9122 | ABL2 | ADCK3 | 0.623529 | 0.470588 | 0.435294 | 0.258824 | 0.235294 | 0.305882 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 9 | 12347 | 1641 | BRAF | CHK1 | 0.346154 | 0.423077 | 0.346154 | 0.243590 | 0.115385 | 0.217949 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 10 | 12347 | 3833 | BRAF | AAK1 | 0.435897 | 0.474359 | 0.333333 | 0.230769 | 0.102564 | 0.230769 | ... | 0.92 | 0.65 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 11 | 12347 | 9122 | BRAF | ADCK3 | 0.576923 | 0.371795 | 0.397436 | 0.256410 | 0.269231 | 0.282051 | ... | 0.92 | 0.68 | 0.89 | 0.92 | 0.92 | 0.92 | 0.92 | 1.0 | 1.0 | 1.0 |
| 12 | 1641 | 3833 | CHK1 | AAK1 | 0.352941 | 0.411765 | 0.352941 | 0.200000 | 0.082353 | 0.235294 | ... | 1.00 | 0.84 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 13 | 1641 | 9122 | CHK1 | ADCK3 | 0.611765 | 0.494118 | 0.541176 | 0.317647 | 0.282353 | 0.317647 | ... | 1.00 | 0.86 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
| 14 | 3833 | 9122 | AAK1 | ADCK3 | 0.611765 | 0.482353 | 0.494118 | 0.282353 | 0.223529 | 0.341176 | ... | 1.00 | 0.87 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.0 | 1.0 | 1.0 |
15 rows × 34 columns
Generate fingerprint distance for all pairwise structures/fingerprints (FingerprintDistanceGenerator)
Input:
FeatureDistancesGeneratorobject (orFingerprintGeneratorobject) and optionally feature weightsOutput:
FingerprintDistanceGeneratorobject
[10]:
from kissim.comparison import FingerprintDistanceGenerator
fingerprint_distance_generator = FingerprintDistanceGenerator.from_feature_distances_generator(
feature_distances_generator
)
Note: Fingerprint distances can also be calculated directy from the fingerprints (FingerprintGenerator > FingerprintDistanceGenerator) instead of using the feature distances explicitly as show above (FingerprintGenerator > FeatureDistancesGenerator > FingerprintDistanceGenerator):
[11]:
fingerprint_distance_generator = FingerprintDistanceGenerator.from_fingerprint_generator(
fingerprint_generator
)
[12]:
fingerprint_distance_generator.data.head()
[12]:
| structure.1 | structure.2 | kinase.1 | kinase.2 | distance | bit_coverage | |
|---|---|---|---|---|---|---|
| 0 | 109 | 118 | ABL2 | ABL2 | 0.074214 | 0.992000 |
| 1 | 109 | 12347 | ABL2 | BRAF | 0.259053 | 0.919333 |
| 2 | 109 | 1641 | ABL2 | CHK1 | 0.253045 | 0.990667 |
| 3 | 109 | 3833 | ABL2 | AAK1 | 0.277368 | 0.990667 |
| 4 | 109 | 9122 | ABL2 | ADCK3 | 0.358882 | 0.990667 |
Kinase distance matrix
[13]:
fingerprint_distance_generator.kinase_distance_matrix(by="minimum")
[13]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | BRAF | CHK1 |
|---|---|---|---|---|---|
| kinase.1 | |||||
| AAK1 | 0.000000 | 0.277368 | 0.303542 | 0.307277 | 0.229590 |
| ABL2 | 0.277368 | 0.000000 | 0.358882 | 0.259053 | 0.246844 |
| ADCK3 | 0.303542 | 0.358882 | 0.000000 | 0.376875 | 0.347142 |
| BRAF | 0.307277 | 0.259053 | 0.376875 | 0.000000 | 0.303330 |
| CHK1 | 0.229590 | 0.246844 | 0.347142 | 0.303330 | 0.000000 |
Show on diagonal experimental values for structure pairs representing each kinase pair (as opposed to simply setting the diagonal to 0 by default).
[14]:
fingerprint_distance_generator.kinase_distance_matrix(by="minimum", fill_diagonal=False)
[14]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | BRAF | CHK1 |
|---|---|---|---|---|---|
| kinase.1 | |||||
| AAK1 | NaN | 0.277368 | 0.303542 | 0.307277 | 0.229590 |
| ABL2 | 0.277368 | 0.074214 | 0.358882 | 0.259053 | 0.246844 |
| ADCK3 | 0.303542 | 0.358882 | NaN | 0.376875 | 0.347142 |
| BRAF | 0.307277 | 0.259053 | 0.376875 | NaN | 0.303330 |
| CHK1 | 0.229590 | 0.246844 | 0.347142 | 0.303330 | NaN |
More structure-kinase mapping methods are available, e.g. maximum or mean. Additionally, the number of structure pairs per kinase pair can be fetched.
[15]:
fingerprint_distance_generator.kinase_distance_matrix(by="size")
[15]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | BRAF | CHK1 |
|---|---|---|---|---|---|
| kinase.1 | |||||
| AAK1 | 0 | 2 | 1 | 1 | 1 |
| ABL2 | 2 | 1 | 2 | 2 | 2 |
| ADCK3 | 1 | 2 | 0 | 1 | 1 |
| BRAF | 1 | 2 | 1 | 0 | 1 |
| CHK1 | 1 | 2 | 1 | 1 | 0 |
[16]:
fingerprint_distance_generator.kinase_distance_matrix(by="std")
[16]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | BRAF | CHK1 |
|---|---|---|---|---|---|
| kinase.1 | |||||
| AAK1 | 0.000 | 0.004 | NaN | NaN | NaN |
| ABL2 | 0.004 | 0.000 | 0.001 | 0.01 | 0.004 |
| ADCK3 | NaN | 0.001 | 0.000 | NaN | NaN |
| BRAF | NaN | 0.010 | NaN | 0.00 | NaN |
| CHK1 | NaN | 0.004 | NaN | NaN | 0.000 |
The kinase distance matrix can also be filtered for kinase pairs with a user-defined bit coverage.
[17]:
fingerprint_distance_generator.kinase_distance_matrix(by="minimum", coverage_min=0.99)
[17]:
| kinase.2 | AAK1 | ABL2 | ADCK3 | CHK1 |
|---|---|---|---|---|
| kinase.1 | ||||
| AAK1 | 0.000000 | 0.277368 | NaN | NaN |
| ABL2 | 0.277368 | 0.000000 | 0.358882 | 0.253045 |
| ADCK3 | NaN | 0.358882 | 0.000000 | NaN |
| CHK1 | NaN | 0.253045 | NaN | 0.000000 |
If you are interested in more information about selected structure pairs (in case of methods minimum and maximum), please use the following method:
[18]:
fingerprint_distance_generator.kinase_distances(by="minimum").head()
[18]:
| index | structure.1 | structure.2 | distance | bit_coverage | ||
|---|---|---|---|---|---|---|
| kinase.1 | kinase.2 | |||||
| ABL2 | ABL2 | 0 | 109 | 118 | 0.074214 | 0.992000 |
| BRAF | 1 | 109 | 12347 | 0.259053 | 0.919333 | |
| CHK1 | 6 | 118 | 1641 | 0.246844 | 0.989333 | |
| AAK1 | 3 | 109 | 3833 | 0.277368 | 0.990667 | |
| ADCK3 | 4 | 109 | 9122 | 0.358882 | 0.990667 |
Structure distance matrix
[19]:
fingerprint_distance_generator.structure_distance_matrix()
[19]:
| structure.2 | 109 | 118 | 1641 | 3833 | 9122 | 12347 |
|---|---|---|---|---|---|---|
| structure.1 | ||||||
| 109 | 0.000000 | 0.074214 | 0.253045 | 0.277368 | 0.358882 | 0.259053 |
| 118 | 0.074214 | 0.000000 | 0.246844 | 0.282949 | 0.360833 | 0.273133 |
| 1641 | 0.253045 | 0.246844 | 0.000000 | 0.229590 | 0.347142 | 0.303330 |
| 3833 | 0.277368 | 0.282949 | 0.229590 | 0.000000 | 0.303542 | 0.307277 |
| 9122 | 0.358882 | 0.360833 | 0.347142 | 0.303542 | 0.000000 | 0.376875 |
| 12347 | 0.259053 | 0.273133 | 0.303330 | 0.307277 | 0.376875 | 0.000000 |
The structure distance matrix can also be filtered for structure pairs with a user-defined bit coverage.
[20]:
fingerprint_distance_generator.structure_distance_matrix(coverage_min=0.99)
[20]:
| structure.2 | 109 | 118 | 1641 | 3833 | 9122 |
|---|---|---|---|---|---|
| structure.1 | |||||
| 109 | 0.000000 | 0.074214 | 0.253045 | 0.277368 | 0.358882 |
| 118 | 0.074214 | 0.000000 | NaN | NaN | NaN |
| 1641 | 0.253045 | NaN | 0.000000 | NaN | NaN |
| 3833 | 0.277368 | NaN | NaN | 0.000000 | NaN |
| 9122 | 0.358882 | NaN | NaN | NaN | 0.000000 |